Protein Expression
Protein Expression and Purification Core Facility at the UofL Health -Brown Cancer Center (CTRB Rm 233)
Core Summary
The Protein Expression and Purification Core provides purified proteins at relatively low cost and minimal investigator effort. We offer recombinant protein expression and purification in E. coli systems to all University of Louisville investigators who need recombinant proteins for functional and structural studies. We also provide specialized technical training and consultation in recombinant protein production. The PI may choose to receive an expression strain, optimized expression and purification scheme, or a purified protein.
Core Details
Typically, our client investigator provides an expression vector for their target protein, however, if needed, we can also design constructs and perform cloning. We use the In-Fusion polymerase chain reaction (PCR) cloning system (Clontech) and mostly pET-based vectors (Novagen) for expression in bacteria. The recombinant clones are transformed into E. coli expression strains. Recombinant protein expression is induced after addition of IPTG in bacteria. We purify tagged (such as HIS, GST, CBD) proteins using ÄKTA Go, Pure, and Purifier (Cytiva) FPLC chromatography systems. The FPLCs are preparative liquid chromatography systems for fast and easy purification of proteins, and for workflow scouting. The use of both instruments allows us to perform affinity, size exclusion and ionic exchange chromatography and complete purification usually on the same day. A typical yield can be as high as 5-10 mg of properly folded material from a liter of bacterial culture. We use thermal melt assays for buffer optimization if there is a sign of protein aggregation/precipitation. The protein target is verified with mass spectrometry (Mass Spectrometry Facility, University of Arkansas) and protein folding can be verified when a sample for Nuclear Magnetic Resonance Spectroscopy (NMR) is produced.
Terms and Conditions
The core support comes from institutional resources, resulting in substantially lower charge-back fees. Publications containing data or products generated by the core require citation in the acknowledgement. Co-authorship is expected for core personnel who provide substantial intellectual input for a project.
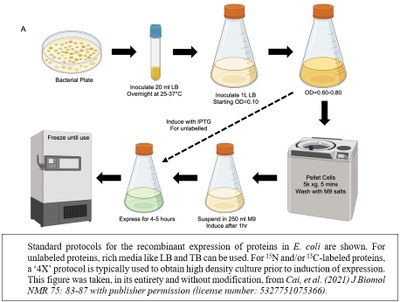
Services
Bacterial protein expression systems are popular because bacteria are easy to culture, grow fast and produce high yields of recombinant protein. We screen a variety of conditions including growth medium, induction temperature, inducer concentration and incubation time to achieve maximum expression of soluble protein in bacteria. Usually we use affinity chromatography, and if necessary, we further purify proteins using ion exchange, hydrophobic and size exclusion chromatography. Regular purification yield is usually in milligram quantities from a liter of culture. We use published, client supplied, or de novo protocols. During our first meeting, the client should explain what the final use of their favorite protein will be in terms of amount, purity, and activity. We will design the strategy to reach the desired goal and present it during the second meeting.
The following is a list of services the Protein Expression Core provides:
- Expression Vector Construction
- Growth Media Preparation
- Transformation
- SDS-PAGE Gel Service
- Expression Optimization
- Large-scale Expression
- Isotopically labeled Protein Expression for NMR
- Cell extract preparation
- Protein Purification
- Protein Refolding
- Tag Removal
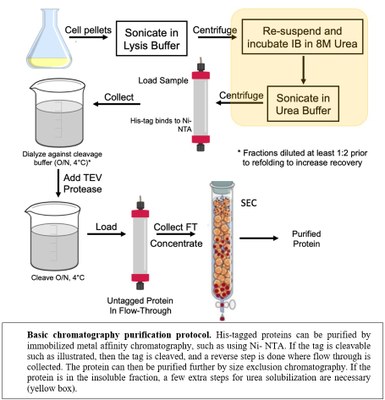
For a new protein system, an estimated timeframe for completion of each task is provided below.
1. Expression vector construction - Gene synthesis by PCR, cloning, plasmid preparation and DNA sequence verification. 2 weeks
2. Transformation into appropriate bacterial expression strain. Protein expression optimization. Protein refolding if necessary. Purification of the protein. Tag-removal if necessary. Product analysis using SDS-PAGE, mass spectrometry, and NMR spectroscopy (if applicable). 2-3 weeks
3. Scale up production, purification, and if necessary, tag removal. 1-2 weeks
Note. It may take more time for challenging proteins, if toxicity to the host, low expression, or low solubility occurs.
Contact Information
Director, Dr. T. Michael Sabo
mike.sabo@louisville.edu tel. (502) 852-3066
Research Associate, Dr. William Holmes
william.holmes@louisville.edu tel. (502) 852-4906
Research Technician II, Anne Carenbauer
anne.carenbauer@louisville.edu tel. (502) 852-4906
 Facebook
Facebook Twitter
Twitter Linkedin
Linkedin