Donald Demuth, PhD

Emeritus - Professor & University Scholar
Emeritus - Associate Dean for Research
Dept Oral Immunology & Infectious Diseases
Summary of Research
- bacterial cell-to-cell communication
- molecular mechanisms of microbial biofilm development
- development of targeted therapeutics for oral pathogens
- nanoparticle drug delivery systems for oral applications
Research Interests:
To colonize and persist in a host, bacterial pathogens must first adhere tightly to tissue surfaces and evade or overcome the host immune response as well as various non-immune anti-microbial activities. Our studies focus on the mechanisms used by pathogens to accomplish these tasks and we utilize the complex microbial community of the human oral cavity as a model to study these processes. Organisms may passively avoid the host immune response by growing as a biofilm. We are interested in determining how the oral pathogen, P. gingivalis identifies a suitable niche and colonizes the complex microbial community that exists in the oral cavity. These early colonization events represent ideal targets for therapeutic intervention and we are currently evaluating both peptide and small molecule compounds that specifically target P. gingivaliscolonization. The development and growth of complex microbial communities may also require communication among the cells within the biofilm and between bacteria and host cells. Therefore, we are investigating mechanisms of bacterial communication, focusing on quorum sensing systems that are present in oral pathogens. The goal of these studies is to determine if quorum sensing plays a role in allowing organisms to sense and respond to their local environment and the other bacteria that cohabit that environment.
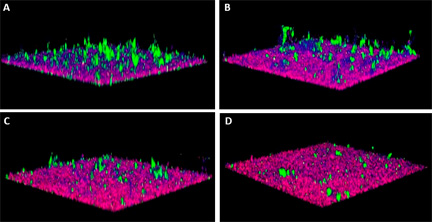
Figure 1. Representative images of a three species biofilm comprising S. gordonii (red), F. nucleatum (blue) and P. gingivalis (green). Control biofilm (A) exposed to buffer alone; (B, C, D) biofilms exposed to buffer containing 5uM, 10uM or 20uM peptidomimetic compound PCP-III, respectively.
Current Projects
1) The periodontal pathogen P. gingivalis colonizes the pre-existing oral biofilm by interacting with a surface protein (SspB) expressed by all oral streptococci. However, P. gingivalis selectively adheres to the viridans streptococci and not mutans streptococci. We have identified a discrete motif of SspB that is involved in this interaction. Using a protein engineering approach, we are investigating the specific protein structure that is recognized by P. gingivalis in order to generate peptide mimetic compounds that may block P. gingivalis colonization of the oral cavity. We are also designing and synthesizing small molecule peptidomimetics that function to reduce or inhibit P. gingivalis colonization of the oral cavity.
2) Drug delivery in the oral cavity is limited by transient exposure times resulting from salivary flow. In collaboration with a bioengineer, we are developing nanoparticle and nanofiber systems that deliver high local doses of active agents in the oral cavity for prolonged periods of time. We are also exploring approaches to target nanoparticles to specific niches in the oral cavity to optimize their drug release characteristics. One method utilizes the Actinomyces coaggregation factor (CafA) to target sustained release nanoparticles to the surface of commensal streptococci, which is the initial niche that is occupied by P. gingivalis.
3) The gene responsible for the production of the auto-inducer 2 signal (AI-2) is conserved in many bacteria, yet whether it plays a role in cell-to-cell communication in all organisms remains controversial. We are interested in determining the mechanisms of cell-to-cell communication mediated by AI-2 in the oral pathogen A. actinomycetemcomitans. We have identified several components are involved in AI-2 signal transduction, e.g., the two component system QseBC, and showed that this systems regulates a large group of genes encoding proteins involved in anaerobic metabolism and other genes involved in iron acquisition. QseBC may also be controlled by toxin-antitoxin (TA) systems and we are exploring the function of several classes of TA systems in A. actinomycetemcomitans.
Recent Publications:
Mahmoud, M.Y., Steinbach-Rankins, J.M.*and Demuth, D.R.* 2019. Functional Assessment of BAR-Modified PLGA Nanoparticles against Oral Biofilms in a Murine Model of Periodontitis. J. Controlled Release. In press. *, corresponding author
Patil, P.C., Tan, J., Demuth, D.R.* and Luzzio, F.A.* 2019. “Second-generation 1,2,3-Triazole-based inhibitors of Porphyromonas gingivalisadherence to oral streptococci and biofilm formation. MedChemComm. DOI: 10.1039/c8md00405f *, corresponding author
Mahmoud, M.Y., Demuth, D.R.* and Steinbach-Rankins, J.M.* 2018. BAR-encapsulated nanoparticles for the inhibition and disruption of Porphyromonas gingivalis- Streptococcus gordoniibiofilms. J. Nanobiotech. 16(1):69. doi: 10.1186/s12951-018-0396-4. *, corresponding author
J. Tan, P.C. Pravin, F.A. Luzzio and D.R. Demuth. 2018. In vitro and in vivo activity of peptidomimetic compounds targeting the periodontal pathogen Porphyromonas gingivalis. Antimicrob. Ag. Chemother. 62(7). pii: e00400-18. doi: 10.1128/AAC.00400-18.
Schneider, B., Weigel, W., Sztukowska, M.N, and Demuth, D.R. 2018. Identification and functional characterization of type II toxin/antitoxin systems in Aggregatibacter actinomycetemcomitans. Molec. Oral Microbiol. 33:224-233.
Kalia, P., Jain, A., Radhakrishnan, R., Demuth, D.R.* and Steinbach-Rankins, J.M.* 2017. Peptide-modified nanoparticles inhibit formation of Porphyromonas gingivalis biofilms with Streptococcus gordonii. Int. J. Nanomed. 12:4553-4562. *, corresponding authors
Chen, S.G., Stribinskis, V., Rane, M.J., Demuth, D.R., Gozal, E., Roberts, A.M., Jagadapillai, R., Liu, R., Choe, K., Shivakumar, B., Son, F., Jin, S., Kerber, R., Adame, A., Masliah A. and Friedland, R.P. 2016. Exposure to the Functional Bacterial Amyloid Protein Curli Enhances Alpha-Synuclein Aggregation in Aged Fischer 344 Rats and Caenorhabditis elegans. Scientific Reports 6:34477. Doi: 10.10038/srep34477.
Patil, P.C., Tan, J., Demuth, D.R.* and Luzzio, F.A.*. 2016. 1,2,3-triazole-based inhibitors of Porphyromonas gingivalisadherence to oral streptococci and biofilm formation. Bioorg Med Chem. 24(21):5410-5417. *, corresponding authors
Weigel, W.A. and Demuth, D.R. 2016. QseBC, a two component bacterial adrenergic receptor and global regulator of virulence in Enterobacteriaceae and Pasteurellaceae. Mol. Oral Microbiol. 31:379-397.
Weigel, W.A., Demuth, D.R.*, Torres Escobar, A. and Juarez-Rodriguez, M.D. 2015. Aggregatibacter actinomycetemcomitans QseBC is activated by catecholamines and iron and regulates genes encoding proteins associated with anaerobic respiration and metabolism. Mol. Oral Microbiol. 30:384-398. *, corresponding author
Patil, P.C., Luzzio, F.A. and Demuth, D.R. 2015. Oxazoles for Click Chemistry II: Synthesis of Extended Heterocyclic Scaffolds. Tetrahedron Letters 56:3039-3041