Since its inception, CREAM has operated using two distinct mechanisms: collaborative research and charge-for-service. In the first approach, the Investigators at CREAM participate in a research proposal by providing their research expertise. If the proposal is awarded, the CREAM budget originally proposed will be used for recharge fees for personnel, routine reagent costs, and minimal instrument fees. For the charge-for-service mechanism, the users are charged based on the service provided by CREAM. To date, CREAM has interacted with more than 60 research groups national wide. Our service has resulted in > 300 peer-reviewed publications, and some of our work also contributed to securing > $250 M of federal research funds.
Mass Spectrometry
The CREAM Mass Spectrometry Facility houses a unique combination of mass spectrometers that can implement various analytical projects and is typically capable of achieving global metabolite and lipid profiling, targeted metabolomics, and stable isotope-resolved metabolomics. Due to the analytical power of each MS and the wide range of applications represented by the combination, many problems outside of metabolomics can also be addressed.
The following provides rates for primary services offered by CREAM, focusing on collecting chemical mass spectra for user samples. However, support is also offered for experimental design, selection of appropriate instrumentation, sample preparation, data interpretation, and appropriate informatics. Samples can be received in various states of preparation for analysis, and clients have options to complete preparation at CREAM and/or pay for preparation.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Notes:
- Sample processing will not be charged if you process samples.
- The cost of constructing the calibration curve will not be charged if CREAM receives more than 10 samples.
- Our 2DLC is configured in parallel mode for profiling polar metabolites and comprehensive mode for profiling lipids using HILIC and RP columns. Each sample is analyzed on the 2DLC-MS system in negative and positive modes to obtain full MS spectra. Group-based pooled samples are also prepared, and 2DLC-MS/MS analyzes each pooled sample to obtain MS/MS spectra.
- All non-UofL customers will be charged an additional 75% cost to cover the F&A. The minimum project cost is $1000.00.
- Use the Instrument Request Form below to request services.
Nuclear Magnetic Resonance
Under construction.
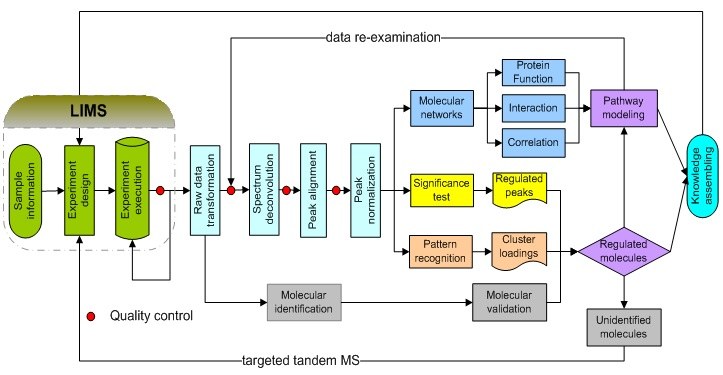
Bioinformatics infrastructure of metabolomics research at the University of Louisville.
Bioinformatics
The bioinformatics research effort at CREAM has resulted in a suite of bioinformatics tools for deciphering biological information from massive metabolomics data, including GC-MS, GC×GC-TOF MS, direct infusion FTICR-MS, LC-MS, NMR. The deconvoluted instrument data are further employed for molecular association network and pathway analyses. These algorithms have been integrated into three data analysis pipelines: PDP, MetSign, and MetPP. In the last few years, we further integrated these pipelines and created a comprehensive bioinformatics infrastructure successfully employed in many biomedical projects.
At the University level, we are supported by an 8-gigabit campus backbone network comprised of over 77 miles of fiber in a dual ring configuration. The network can provide 10 Mbps, 100 Mbps, and 1 Gbps Ethernet service for faculty and staff communications needs. The campus wireless network utilizes 54 Mbps (802.11g) technology and is widely available across the campuses, with access to scheduled classrooms and public areas of most buildings. In addition, the University of Louisville is connected to the Internet2 node via dedicated fiber and currently has access to 1 Gbps of bandwidth. The Internet2 connection gives the University of Louisville direct access to the national research and education networks and enables participation in national networking initiatives such as caGRID/caBIG and the TeraGrid. We use this cluster for bioinformatics development and analyze the experimental data acquired from large-scale metabolomics studies.
