Richard J. Lamont, PhD

Delta Dental Endowed Professor
Chair, Oral Immunology and Infectious Diseases
Director, Center for Microbiomics, Inflammation and Pathogenicity
Room 221B, Baxter I
Phone: 502 852 2112
email:rich.lamont@louisville.edu
Research in the Lamont laboratory is centered on the concept that polymicrobial communities are the fundamental etiological unit in periodontal disease. Indeed, the primary function of what is thought of as a conventional pathogen, Porphyromonas gingivalis, may be to induce a transient and localized immune suppression which allows the outgrowth of pathobionts in periodontal microbial communities. Further, the immunosuppressive properties of P. gingivalis are dependent on the contextual cues provided by co-habiting organisms. It is against this overall backdrop that we are engaged in two main projects with the overall goal of integrating the emergent properties of microbial communities with homeostatic or dysbiotic host responses.
Molecular and cellular dialog between Porphyromonas gingivalis and gingival epithelial cells.
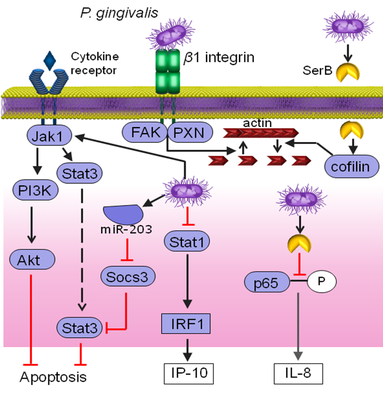
Gingival epithelial cells (GECs) are not simply a passive barrier to microbial intrusion, but continually surveil the subgingival microbiome, and through a network of signaling systems coordinate expression of cytokines and chemokines which direct the recruitment and activation of immune cells. At this interactive interface P. gingivalis impinges upon epithelial signal transduction pathways, many of which are based on patterns of protein phosphorylation and dephosphorylation. Consequently, P. gingivalis can suppress production of: the neutrophil chemokine IL-8; the Th1/Th17 biasing chemokine IP-10; and the antiviral IFN-λ. Additionally, P. gingivalis can suppress epithelial apoptotic cell death which may have relevance for oral squamous cell carcinoma (OSCC).
 |  |
|---|
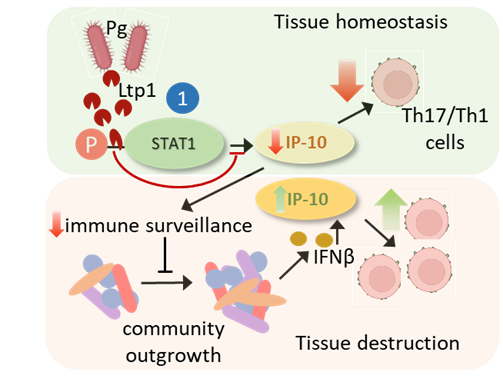
A current focus is on IP-10 which is transcriptionally suppressed through the action of a secreted tyrosine phosphatase Ltp1 which dephosphorylates STAT1. IP-10 promotes gingival immune surveillance, and suppression of IP-10 will thus facilitate community expansion, with eventual reversal of immune suppression and mobilization of Th1 and Th17 cells which are major contributors to gingival tissue destruction.
Community dependent pathogenicity of P. gingivalis
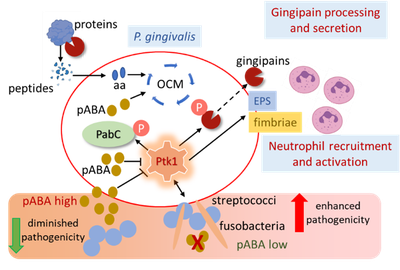
The pathophysiology of P. gingivalis is intricately interconnected with the multispecies community in which it resides. Community pathogenic potential is shaped by complex and dynamic interspecies interactions based on both diffusible signaling molecules and physical association. Emergent properties of pathogenic communities include induction of dysbiotic inflammatory responses which depend to a significant degree on the extent of neutrophil recruitment and activation. We have found that the pathogenicity of P. gingivalis can be constrained by a streptococcal diffusible metabolite, para-amino benzoic acid (pABA), a precursor of de novo folate synthesis in the physiologically essential one-carbon metabolism (OCM) pathway. However, as physically integrated communities develop, interspecies co-adhesion results in suppression of streptococcal pABA production, and partner organisms such as Streptococcus gordonii and Fusobacterium nucleatum enhance overall community pathogenicity. Sensing of both pABA and interspecies physical contact by P. gingivalis funnels through a protein tyrosine phosphorylation signaling circuit. The tyrosine kinase Ptk1 controls flux through OCM as well as the production of virulence factors, including the gingipain proteases, fimbriae and extracellular polysaccharide (EPS). Our current research is directed toward understanding the role of Ptk1 as a master regulator of P. gingivalis virulence which can integrate metabolic and physical cues from the community environment and calibrate the pathophysiological response.
Representative publications
- Hajishengallis G, Lamont RJ, Koo H. 2023. Oral polymicrobial communities: Assembly, function, and impact on diseases. Cell Host Microbe 31:528-538.
- Pandey SD, Perpich JD, Stocke KS, Mansfield JM, Kikuchi Y, Yakoumatos L, Muszyński A, Azadi P, Tettelin H, Whiteley M, Uriarte SM, Bagaitkar J, Vickerman M, Lamont RJ. 2023. Impact of polymicrobial infection on fitness of Streptococcus gordonii in vivo. mBio 14:e0065823.
- Song L, Perpich JD, Wu C, Doan T, Nowakowska Z, Potempa J, Christie PJ, Cascales E, Lamont RJ, Hu B. 2022. A unique bacterial secretion machinery with multiple secretion centers. Proc Natl Acad Sci U S A 119:e2119907119.
- Lewin GR, Stocke KS, Lamont RJ, Whiteley M. 2022. A quantitative framework reveals traditional laboratory growth is a highly accurate model of human oral infection. Proc Natl Acad Sci U S A 119:e2116637119.
- Rodriguez-Hernandez CJ, Sokoloski KJ, Stocke KS, Dukka H, Jin S, Metzler MA, Zaitsev K, Shpak B, Shen D, Miller DP, Artyomov MN, Lamont RJ, Bagaitkar J. 2021. Microbiome-mediated incapacitation of interferon lambda production in the oral mucosa. Proc Natl Acad Sci U S A 118:e2105170118.
- Fitzsimonds ZR, Liu C, Stocke KS, Yakoumatos L, Shumway B, Miller DP, Artyomov MN, Bagaitkar J, Lamont RJ. 2021. Regulation of olfactomedin 4 by Porphyromonas gingivalis in a community context. ISME J 15:2627-2642.
- Jung YJ, Miller DP, Perpich JD, Fitzsimonds ZR, Shen D, Ohshima J, Lamont RJ. 2019. Porphyromonas gingivalis tyrosine phosphatase Php1 promotes community development and pathogenicity. mBio 10:e02004.
- Lewin GR, Stacy A, Michie KL, Lamont RJ, Whiteley M. 2019. Large-scale identification of pathogen essential genes during coinfection with sympatric and allopatric microbes. Proc Natl Acad Sci U S A 116:19685-19694.
- Ohshima J, Wang Q, Fitzsimonds ZR, Miller DP, Sztukowska MN, Jung YJ, Hayashi M, Whiteley M, Lamont RJ. 2019. Streptococcus gordonii programs epithelial cells to resist ZEB2 induction by Porphyromonas gingivalis. Proc Natl Acad Sci U S A 116:8535-8543.
- Lamont RJ, Koo H, Hajishengallis GH. 2018. The oral microbiota: dynamic communities and host interactions. Nature Rev Microbiol 16:745-759.
- Kuboniwa M, Houser JR, Hendrickson EL, Wang Q, Alghamdi SA, Sakanaka A, Miller DP, Hutcherson JA, Wang T, Beck DAC, Whiteley M, Amano A, Wang H, Marcotte EM, Hackett M, Lamont RJ. 2017. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat Microbiol. 2: 1493-1499.
- Tribble GD, Mao S, James CE, Lamont RJ. 2006. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc Natl Acad Sci U S A 103:11027-11032.
Complete List of Published Work in MyBibliography